What’s new in e!75?
- New VEP interface
- New “Age of base” track for human
- A link to individual exons is now available in the Region in Detail page
- New Gencode Basic Gene Set renderer for human and mouse
New VEP interface
Ensembl has a number of data tools that can benefit from offline processing to overcome the time-out limits imposed by the web. We have therefore created a new interface where you can submit data to these tools and view all your tickets: the first to be ported is the VEP, with others planned for future releases.
The new VEP interface will allow you to process many more variants and is shipped with a new job page where you can easily monitor all your recent tickets, edit, remove or save them into your account. More details on the input form and job page can be found on the VEP Input page.
The result page has been greatly improved, various statistics are now listed in a table
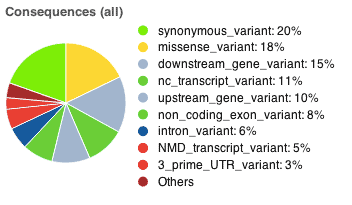 including lines of input read, variants processed and variants remaining after filtering. The result page also includes pie charts showing detailed proportions of consequence types called across all variants. The page now allows you to export the result table in VCF, VEP and TXT format. More details about the results can be found on the VEP Results page.
including lines of input read, variants processed and variants remaining after filtering. The result page also includes pie charts showing detailed proportions of consequence types called across all variants. The page now allows you to export the result table in VCF, VEP and TXT format. More details about the results can be found on the VEP Results page.
New “Age of base” track for human
We have added a new track for human, showing the timing of the most recent mutations as determined by inter-species whole genome alignments. You can find the track in the comparative genomics menu under “Conservation regions” (or search for “age of base”).
Each base pair in which the human reference genome differs by substitution from one of its inferred ancestral genomes is coloured in either grey (event prior to the primate branch), blue (primate specific), red (human-specific, fixed variant), or yellow (human-specific segregating variant, i.e. SNP). Clicking on a mutation position reveals the sub-tree of species which have inherited the same mutation from their common ancestor. It also reveals a score that represents the age of the mutation in arbitrary units, and determines the intensity of the colouring. The more recent the mutation, the lower the score and the darker the colour.
Note that this is a beta version of the track – if you find it useful, please let us know!
Individual exon link in the Region in Detail
The popup menu that appears when you click on a transcript now includes a link to the Exon table. The Exon ID from this popup will be displayed in bold on the Exon page.
Note that this link will only appear when you are zoomed in enough for the click coordinates to clearly identify a single exon.
GENCODE Basic Gene Set Renderer
You can now view the GENCODE gene set in a new way, called “GENCODE Basic”. The new GENCODE Basic display option shows only a only a subset of the GENCODE transcripts.
When using the GENCODE Basic display option, partial transcripts will not be shown if the gene includes complete (Met to Stop) transcripts. Problematic biotypes (eg. retained intron) are excluded from the basic set.
The full GENCODE gene set will be available as before, using the standard display options such as “Expanded with labels”.
Other news:
- Updated zebrafish gene set to include manual annotation from Havana
- Added RNA-seq data for mouse
- Added variation data for turkey from dbSNP build 139
- Imported the latest sequence variants from dbSNP build 139 for dog, horse, opossum, platypus and zebra finch.
- Imported mouse phenotype data from IMPC (International Mouse Phenotyping Consortium)
- Updated variation citations from EPMC and UCSC
- Highlighting of the current feature can now be disabled
- Updated Caenorhabditis elegans reference annotation to Wormbase WS240
- Renamed Saccharomyces cerevisiae assembly from EF4 to R64-1-1 and liftover mapping added
A complete list of the changes can be found on the Ensembl website.
Find out more at the Ensembl Release Webinar e75 (Wed, Mar 12, 2014 4:00 PM – 4:20 PM GMT). Register for free here: http://tinyurl.com/e75webinar










 Assembly patches will be added and annotated for human (GRCh37.p13) and mouse (GRCm38.p2)
Assembly patches will be added and annotated for human (GRCh37.p13) and mouse (GRCm38.p2)







 SIFT scores for all possible changes to the proteome in human, mouse, zebrafish, pig, cow, chicken, rat and dog
SIFT scores for all possible changes to the proteome in human, mouse, zebrafish, pig, cow, chicken, rat and dog




