What’s new in e83:
- Human: gene set updated to GENCODE 24, and new assembly patches (GRCh38.p5)
- Manhattan plot track for LD
- Advanced Filtering and Counts on Variant table
- Minor allele frequency (MAF) filter on sequence mark-up views
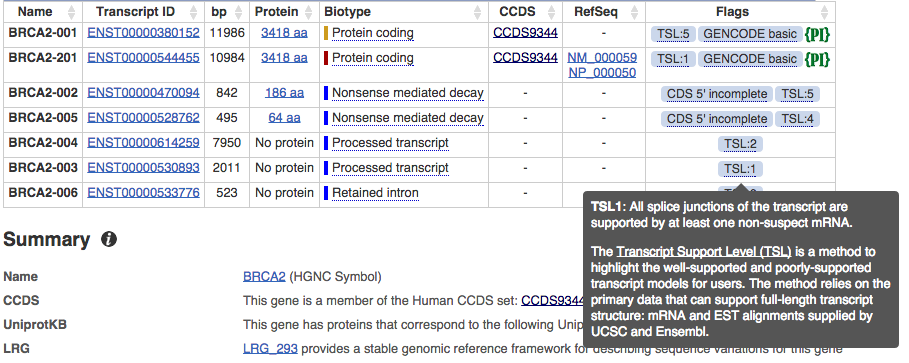
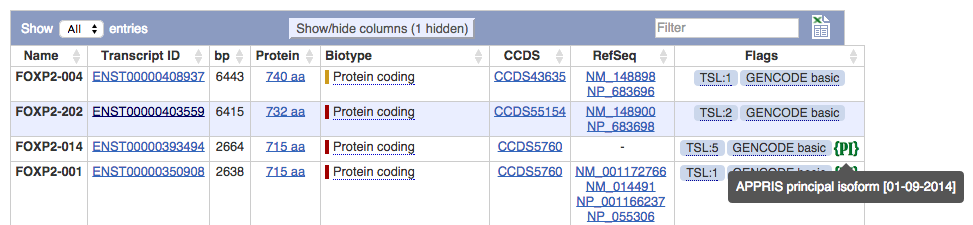
Human gene set update and new assembly patches
The human gene set now corresponds to GENCODE 24 while the assembly has been updated to include new assembly patches for GRCh38.p5.
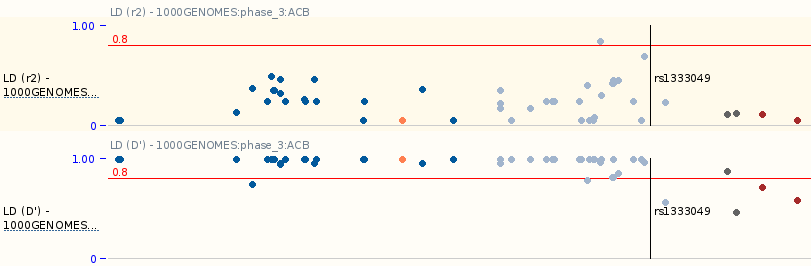
Manhattan plot track for linkage disequilibrium
This new linkage disequilibrium (LD) track is focused on a variant and displays the linked variants surrounding the focus variant. The track displays a Manhattan plot, using the r2 and D prime values (from 0 to 1) on the Y axis. The new track is accessible in the Variation Linkage disequilibrium page, through the links in the new column “LD Manhattan plot“.
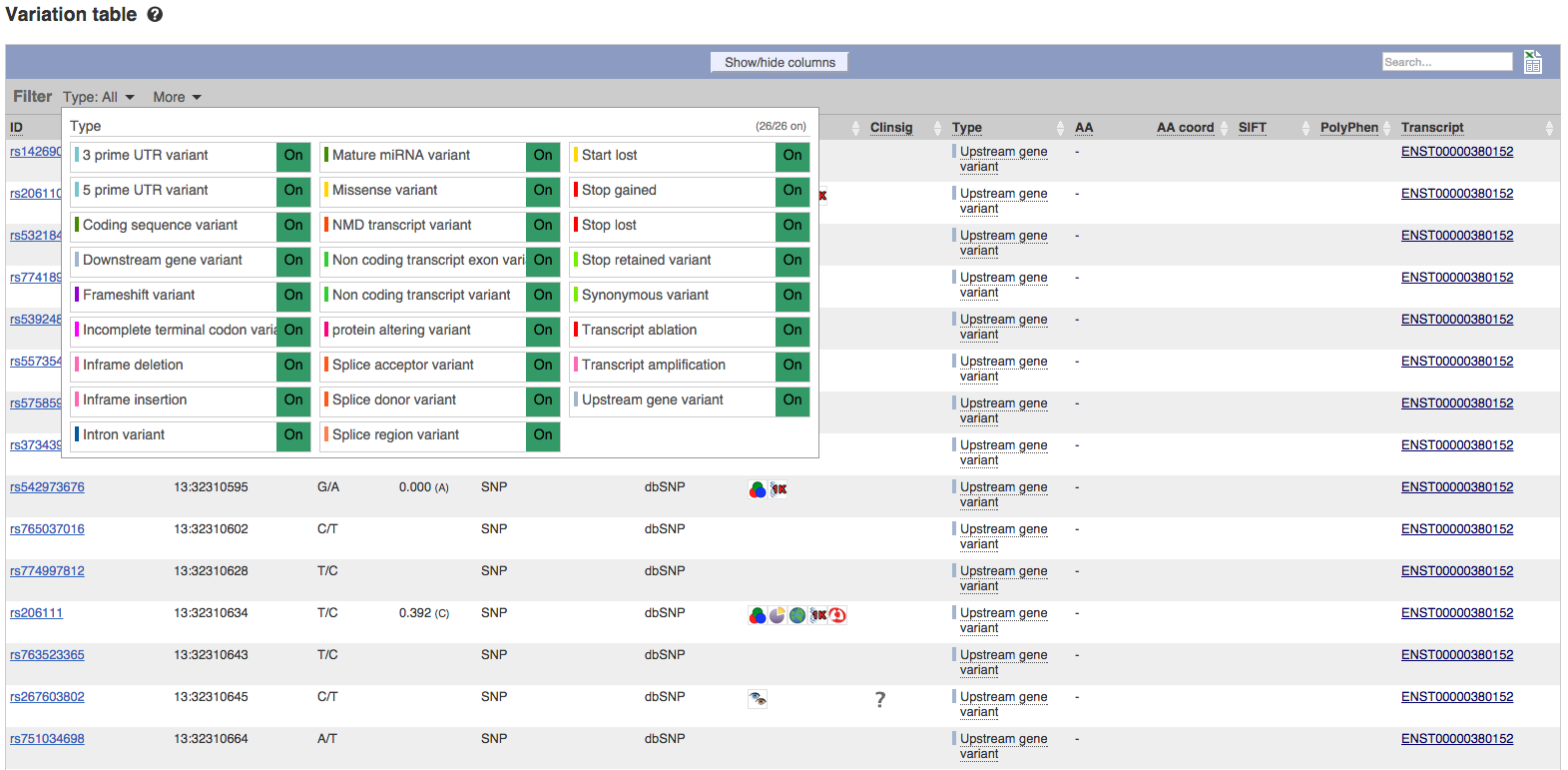
Advanced Filtering and Counts on Variant table
The functionality of the variant table has been further expanded to allow a wider range of filtering options. Filtering can now be applied by Minor Allele Frequency, SIFT and PolyPhen scores, Clinical Significance, Consequence Type and many other columns, using buttons along the top of the variant table. For many of these filters, preset useful combinations of options are available within the popup allowing rapid configuration of more complex combinations. In addition, row counts for each consequence type have been readded to the existing Consequence Type filter. These are displayed in the popup which appears once the filter button has been pressed.
MAF filter on sequence mark-up views
The variants displayed on all sequence mark-up views can be filtered by minor allele frequency (MAF), allowing you to either show or hide according to a range of frequencies (between 0.01% and 10%). This filtering is not on by default so to enable it go to ‘Configure this page’ on any sequence view page and then choose the value you want from the ‘Hide variants by frequency (MAF)’ drop down menu.
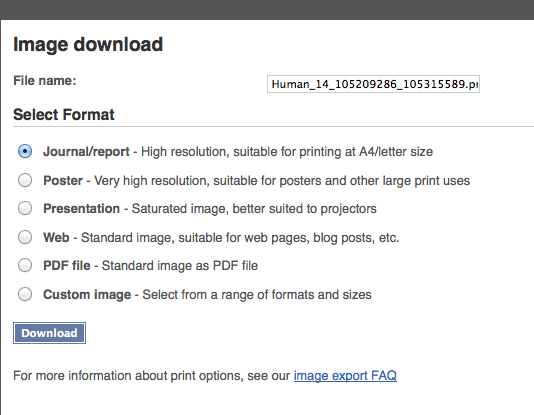
 Improving the image export and Ensembl mobile website
Improving the image export and Ensembl mobile website
There are many updates to these functionalities which will be described in detail in separate blog posts. Look out for these blogs! Below is how the new image export wizard looks like.
Other news
- Mouse: updated to GENCODE M8 annotation
- Rat: updated gene set, including manually annotated HAVANA annotation
- Annotations now available in RDF format for all species on our FTP site
- New human phenotype association data from Cancer Gene Census
- RefSeq genomic to mRNA comparison attributes will be updated for human
- New dbSNP145 variation data for chicken and pig
A complete list of the changes can be found on the Ensembl website.
Find out more about the new release, and ask the team questions, in our free webinar. Wednesday 16th December, 4pm GMT. Register here.