Our initial annotation of the new human assembly is live in the Ensembl Pre site and we are now fully annotating GRCh38 as part of our customary Ensembl release cycle. The complete annotation will be available in Ensembl release 76, due in the third quarter of 2014.
What is a release cycle in Ensembl?
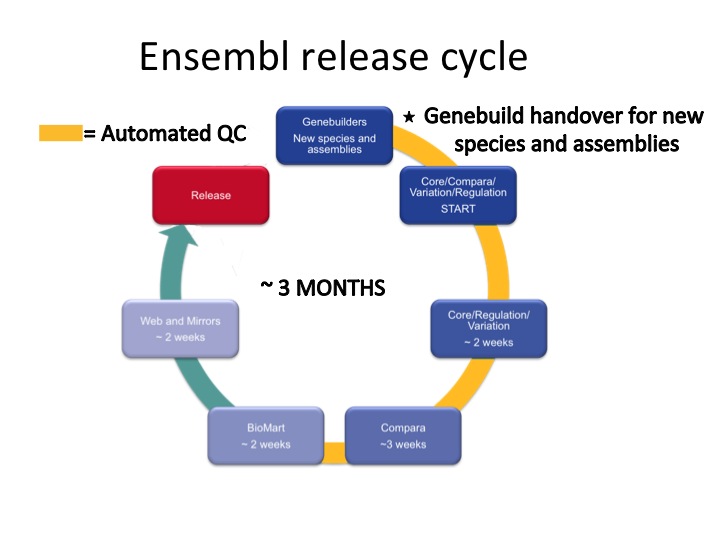
In a nutshell, it’s the process of updating and releasing the Ensembl website and the underlying databases. It takes three months and allows us to provide our data and tools and to perform rigorous steps of quality checking throughout.
A new Ensembl release may include new species, new assemblies, updated gene sets, new variation data, new gene trees, genomic alignments and homologies, and annotation of regulatory features. There will also be improvements and additions to our web-interface and database structure, including the Perl APIs.
Different teams in Ensembl get involved in this orchestrated release cycle. More than 40 people take part in this whole process.

Genebuild
The Genebuild team have developed a gene annotation pipeline to annotate gene and transcripts on a given assembly, based on biological evidence (e.g. protein and nucleotide sequences). This step can take three to six months depending on the quality of the assembly, the number of species-specific sequences available in public sequence databases, and the amount of RNASeq data available, just to name a few. The genebuild team also annotate patches and haplotypes.
For human, mouse and zebrafish, the Ensembl automatic annotation is combined with the Havana manual annotation to give the merged gene set. For human and mouse, these merged genes are the GENCODE gene set. Once these databases are complete, they are handed over to the other Ensembl teams for downstream processing and analyses. The handing over of the databases corresponds to the first step in our release cycle.
Core
The Core team provide the API support for the core and core-like (i.e. otherfeatures, cdna and rnaseq) databases in Ensembl. They also maintain our automated quality control (QC) system in addition to the key pieces of Perl code, such as assembly mapping, Ensembl ID mapping, cross-referencing of Ensembl models to external data sources, such as HGNC, UniProt-KB, RefSeq and GO.
Compara
The Comparative genomics team (also known as Compara) run multiple pipelines to bring the separate species together and provide gene trees (coding and ncRNA), homologues and protein families, in addition to whole genome alignments and synteny data. The resultant data is compiled into the ensembl_compara and ensembl_ancestral databases.
Variation
The Variation team bring together sequence variation data from a variety of sources, predict effect of these variants on our transcripts, call new variants from re-sequencing data, import QTLs, maintain the VEP and incorporate associated disease and phenotype information. These data are used to create variation databases, currently for 22 Ensembl species.
Regulation
The Regulation team collect experimental data from Ensembl collaborators and integrate these data into our regulatory build for human and mouse. This results in potential promoters, enhancers and other regulatory features based on ChIP-seq, DNase I, methylation and other data. Currently we compute a regulatory build for human and mouse. Regulation (also known as funcgen) databases exist for some other species, such as chicken, to support the microarray mapping data. Other sources of regulation can be found on our help page.
Production
Once the Compara, Core, Variation and Regulation teams have handed over their databases, the Production team do a full rebuild of all BioMart databases from the updated data. These data can be accessed via the Ensembl BioMart data-mining tool, martservice, or using the biomaRt package from Bioconductor. The Production team are also responsible for overseeing the entire release cycle and running various pipelines throughout the process to generate statistics for the website, dump flat-files for the FTP site, generate BLAST indices, and carry out quality control of the data.
Web
Whilst the genomic data is being prepared, the Web team work on new displays and website features, maintain BLAST/BLAT in addition to revamping extant online tools, such as our popular VEP. They bring together all the finished databases and make the content available on the Ensembl web browser in a number of ways:
- The website configuration is updated to access the new data
- The databases are copied to the public MySQL servers and dumped for downloading from FTP site.
When the new release is ready to go live, a copy of the current version is set up as an archive, and the web server is updated to point to the new site. A new release also includes updated APIs from many of the teams and tools to access the fully integrated data.
Outreach
The Outreach team carry out usability testing of the new displays and website features developed during the release cycle. The team also update the help and documentation pages, provide user support and online training, deliver workshops and engage with our users via YouTube, Twitter and Facebook. It’s the bridge between the different Ensembl teams and our user communities, so that our users know how and when to access and view the data in our integrated tools and resources.
The special release cycle for GRCh38
 The new assembly of our most popular genome, human, was released by GRC in December 2013.
The new assembly of our most popular genome, human, was released by GRC in December 2013.
Getting a new human assembly requires extra work during the release cycle. In addition to the usual steps described above, we also need to remap and reprocess our data against the new genome. This invariably leads to a longer release cycle.
We want to deliver a top-quality annotation of genes, sequence variants, regulatory elements and comparative features for the new human assembly.
The blog posts from the Ensembl teams in this GRCh38 series will describe the extra steps involved in preparing the updated data in the new human assembly. For more details and comments, just in get in touch.



 Ensembl release 69 is scheduled for release in October.
Ensembl release 69 is scheduled for release in October.






 Confused about browser navigation? Why not try our new elearning course!
Confused about browser navigation? Why not try our new elearning course!