Please note that eg34 is a maintenance release, consisting of fixes to data and software issues. No significant updates have been included.
You can view the release notes here.
 Ensembl Blog
Ensembl BlogPlease note that eg34 is a maintenance release, consisting of fixes to data and software issues. No significant updates have been included.
You can view the release notes here.
In Ensembl 87, there are a number of updates to the assemblies and gene sets for several species:
e87 includes an updated Ensembl-Havana mouse gene set, a merge of complete Ensembl gene models and the latest Havana gene annotation. All CCDS genes are included in this gene set.
This latest Havana gene annotation includes improved gene models for the mouse olfactory receptors. Over 2Mbp of additional sequence has been added to the mouse olfactory genes to create several hundred multi-exonic models. These new models are based on RNA-seq data from Ibarra-Soria X et. al.
The mouse assembly has been updated to GRCm38.p5. The patches for GRCm38.p5 were annotated using a combination of manual annotation, annotation projected from the primary assembly and annotation derived from cDNA and protein alignment evidence.
Due to the high number of epigenomes now available in the Human Regulatory Build, we can no longer show them all by default on the Regulation Summary image, in the Regulation Tab. We have therefore added a table listing the cell types by their regulatory feature activity.
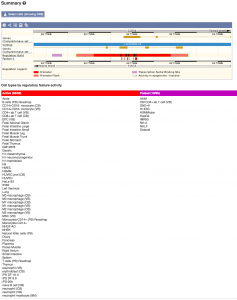
A complete list of the changes can be found on the Ensembl website.
Find out more about the new release and ask the team questions, in our free webinar: Wednesday 14th December, 4pm GMT. Register here.
As part of Ensembl 85, we are excited to introduce expression quantitative trait loci (eQTL) data, through our partnership with the Genotype-Tissue Expression (GTEx) project.
The GTEx project has the goal of identifying the influence of genetics on tissue-specific gene expression, i.e. to map correlations between genotype (SNPs) and gene expression levels (RNA-seq). eQTLs are variants which are found to be significantly correlated with differences in gene expression. Though still in its infancy, we hope that in time this type of data will allow us to conclusively determine the link between regulatory features and their gene targets.
Thanks to our use of HDF5 technology, we offer the only rapid look-up service across all GTEx SNP-gene association tests. We have included all of the correlated variants, including those that fall short of the significance threshold. The GTEx V6 dataset represents 7051 tissue samples from 44 tissues of 449 donors, and a total of 6 billion data points.
To view GTEx eQTL data for any gene, navigate to the gene tab and select ‘regulation’ in the left panel. The display will show one example track of GTEx data for a single tissue. Configuring the page allows you to add more GTEx tracks for each tissue type, by selecting ‘other regulatory regions’ and choosing the tissues you are interested in:
The SNPs are displayed in a Manhattan plot on these tracks, and are coloured according to their consequences on the transcript – as determined by the VEP. Clicking on any of the variants will display correlation statistics and a link to the variant tab. Where the SNPs are clustered, clicking will bring up a list of all variants nearby:
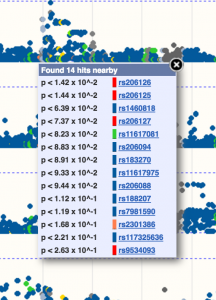
We have also provided Ensembl REST API endpoints to access these data. Currently, these methods allow you to quickly find the beta correlations and their p-values filtered by gene, SNP and/or tissue. You can also list all the tissue types that are currently available on our server.
Currently we are displaying the variants around a gene and their correlation to its expression level. In our next release (e86), on the Variant view, we will display all the genes whose expression levels are correlated to that variant. We will also display the beta effect sizes on the Manhattan plots.
If you have any feedback or questions relating to eQTLs in Ensembl, please contact the helpdesk.
Ensembl Plants now has an archive site, where we will keep selected previous releases of Ensembl Plants publicly available. The first release available on the archive site is release 31, and includes the previous assemblies for wheat and maize.
New assemblies in Ensembl Plants include:
Genome assemblies for 5 new species, including Beta vulgaris (sugar beet), Brassica napus (rapeseed) and Trifolium pratense (red clover)
Ensembl Metazoa: Rfam covariance models have been applied to all metazoan genomes, and are shown in the ‘Rfam models’ track in the genome browser. Click on a model to see the description and the secondary structure.
Ensembl Bacteria now includes the latest versions of 41,610 genomes (41,198 bacteria and 412 archaea) from the INSDC archives. In this release we added 2269 new genomes, 15 genomes with updated assemblies, 212 genomes with updated annotation, 906 genomes where the assigned name has changed, and 243 genomes removed since the last release.
Ensembl Fungi has been updated with 47 newly available genomes and now includes 634 genomes from 388 species. PHI-base references have been added where available, as have non-coding RNA matches to Rfam.
25 new genomes have been added to Ensembl Protists, which now includes 178 genomes from 114 species.
You can find more details in the release notes.
Roadmap Epigenomics is producing epigenomic maps for stem cells and primary ex vivo tissues selected to represent the normal counterparts of tissues and organ systems frequently involved in human disease. In Ensembl 85, we have run our regulatory pipeline on Roadmap Epigenomics data for 30 cell/tissue types.
Additionally, the peak calling component of the Ensembl Regulation Sequencing Analysis pipeline has been improved. All of the existing ENCODE and BLUEPRINT data in Ensembl’s Regulation database have been reprocessed.
The human gene set now corresponds to GENCODE 25 and the assembly has been updated to include new assembly patches for GRCH38.p7
We now import symbol names from the Vertebrate Gene Nomenclature Committee (VGNC) for Chimpanzee. VGNC is an extension of HGNC for standardising naming across vertebrates lacking a nomenclature committee by transferring gene symbols from human to known orthologues. This replaces our own system for naming chimpanzee genes. This has improved our naming of chimpanzee genes as seen in POMGNT2, which was previously named GTDC2 in Ensembl 84.
We have updated the highlighting functions in Location View:
Wasabi has replaced Jalview as a way to view gene trees and multiple alignments. Clicking on any node within the gene tree will give you the option to ‘View in Wasabi’.
Clicking on ‘View in Wasabi’ will open a pop-up window with the tree and alignment:
New tool: File Chameleon customises files from the Ensembl FTP server. Current functions include; adding ‘chr’ to your chromosome names for use with UCSC’s genome browser and removing long genes
A complete list of the changes can be found on the Ensembl website.
Find out more about the new release and ask the team questions, in our free webinar: Wednesday 27th July, 4pm BST. Register here.
Ensembl 85 is scheduled for July 2016, highlights include:
Armadillo/Dog/Ferret: new lincRNA models
For more details on the declared intentions, please visit our Ensembl admin site. Please note that these are intentions and are not guaranteed to make it into the release.
We have scheduled the next releases of Ensembl (Release 85) and Ensembl Genomes (Release 32) for July 2016. Details of the declared intentions will be announced nearer the time.
Please contact the helpdesk if you have any questions or feedback.
We released the new Ensembl mobile site in release 82 and it’s already being used by our communities. m.ensembl.org has a slim-line functionality, designed to be a quick reference tool for genes, variants and associated phenotypes.
We hope you will find this useful if, for example, you’re on your mobile device at a conference, or you’re having coffee with a colleague and want to quickly look up something.
Each page features a share button ![]() , which gives you the option of posting the page to Facebook or Twitter, or emailing the URL to a colleague or even yourself so that you can explore the annotation in more detail on the desktop website when you get back to your computer.
, which gives you the option of posting the page to Facebook or Twitter, or emailing the URL to a colleague or even yourself so that you can explore the annotation in more detail on the desktop website when you get back to your computer.
Next time you are on the go, try m.ensembl.org out! If you have any feedback, please email helpdesk@ensembl.org to share your thoughts with us.