In our latest release of Ensembl, we launched a brand new web interface for the VEP (Variant Effect Predictor).

As “it says on the tin”, the VEP predicts the effect of variants (i.e. SNPs, indels and CNVs) on genes and regulatory elements. It tells you where your variants are located (e.g. introns, coding exons, transcription factor binding motifs), what effect they may have on protein coding sequences, and whether these effects might be deleterious or benign.
The VEP does this by mapping your variants against genes, transcripts, translations, and regulatory features that we annotate in Ensembl.
The Variant Effect Predictor can also be run against other gene sets: you can predict the effect of your variants on RefSeq genes too!
What is new?
The new web interface is more user-friendly and has lots of improvements:
Increased number of variants you can input
You can upload up to one million variants in a compressed format, with a 50MB file size limit. To upload these larger files, you simply need to log in. If you do not have an Ensembl account, you are missing out, as there are many perks of registering. It’s easy to do: just provide your name and email address. If you’d rather not register, the upload limit drops down to 5 MB, i.e. around 100,000 variants.
Display of results
We provide a summary statistics table and pie charts illustrating the different SO terms and the classes of coding consequences for the variants you input.
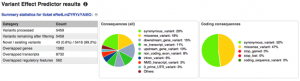
The new web interface provides user-friendly pie charts and summary statistics.
The results preview table with additional details is shown after the pie charts. You can apply a range of filters to any of the data fields and limit the results you see. The full or filtered results can be downloaded as VCF or tab-delimited text for import into Excel.
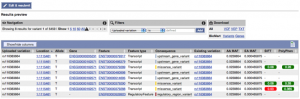
The new ticket tracker
You can run several jobs at the same time and track them back at a later date via the ticket numbers assigned to them. You can easily edit and re-run previous jobs. These jobs will be kept in our Ensembl servers for 30 days. If you register though, the jobs will be kept for as long as you like.

Population frequency data for the 1000 Genomes and ESP projects.
The VEP provides frequency data for known variants from both the 1000 Genomes and NHLBI exome sequencing projects.
You can also use this frequency data to filter your variants: you may wish to exclude known variants with a frequency above 1%, for example.
VEP results are linked to BioMart
The results table in the VEP is now directly linked to BioMart, a data export tool.
This allows you to retrieve additional data about known variants or the genes your variants affect.

You just need to select the attributes in BioMart, e.g. phenotype, orthologues, Gene Ontology terms, and you are ready to go.
Other ways to access the VEP
If you use a command line, you can run the VEP with our script on your own computer. With the Perl script, you can do everything you can do in the online version plus much, much more! It’s the most powerful way to use the VEP.
A couple of functionalities of the VEP (e.g. fetch variant consequences) are also available in the beta version of our language agonistic Rest API.
Help on the VEP
Have a look at our video on the new online VEP interface and our documentation pages for help on the web interface and script versions of VEP.
If you have questions or comments, please get in touch with us.




























