What is new?
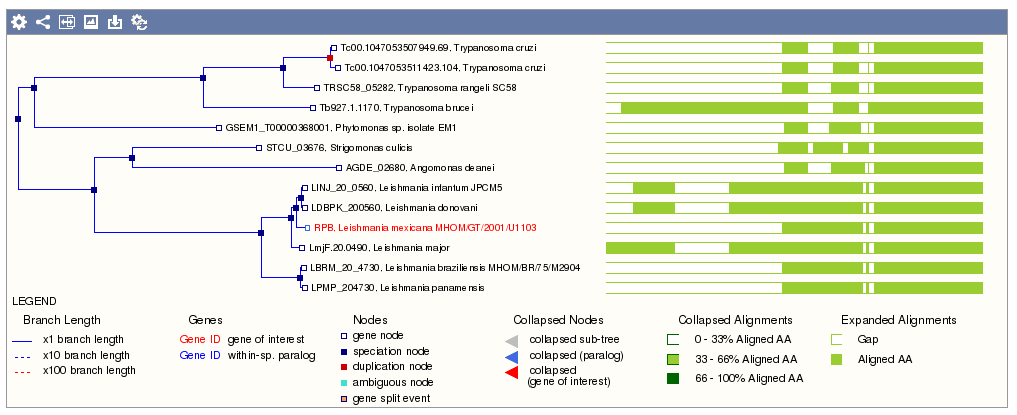
- Molecular and biological information from PHI-base (version 4.0) for thousands of genes in Protists, Bacteria and Fungi that are involved in pathogen-host interactions
- Small non-coding RNA genes in the diatom Phaeodactylum tricornutum (Rogato et al. 2014)

Small non-coding RNA genes described by Rogato et al (2014) in the diatom P. tricornutum are now available in Ensembl Protists. Hover over the ncRNA genes track (genes coloured in light purple) for more information.
- Software update: version 83 of the Ensembl project for all our 30,586 genomes from 5,319 species
Other news
- Improved image export functionality from our websites
Check our ‘New image export option in Ensembl‘ post for more details.

The images in Ensembl Genomes browser websites can be exported in different formats and resolution. Choose the one that suits you best and click on ‘Download’.
- Protein domains for Protists, Metazoa, Fungi and Plants recalculated with InterProScan (version 54.0)
- Updated BioMart: Protists, Metazoa, Fungi and Plants
- Updated gene trees in Plants
Check out all the changes on our Ensembl Genomes website.
Any questions or comments? Email us.