What’s new in e78:
- Instant VEP: immediate answers for a single variant (All species)
- Mouse: genes updated to GENCODE M4, and new assembly patches (GRCm38.p3).
- Transcript Support Levels (TSLs), APPRIS principal isoforms and GENCODE basic annotations are now accessible through Ensembl BioMart (Human, mouse, rat and zebrafish).
- Find Gene Alleles across patches and haplotypes (Human)
- Interactive species tree (All species)
- Improved Gene Tree export (All species)
Instant VEP

It is now possible to get instant VEP results for single variant queries; simply paste a variant in any format into the box and click the button. For more detailed results you can still submit the request in the usual manner.
Mouse update
The mouse genome assembly has been updated to include new assembly patches for GRCm38.p3. We are now displaying GENCODE M4 in Ensembl.

This new assembly patch, KB469738.2 (MG132_PATCH), provides a fix for chromosome 9 for genes Nlrp4g and Ppp2r3d.
APPRIS flags for human, mouse, rat and zebrafish

We have updated the APPRIS flags for human and mouse and also imported the data for rat and zebrafish. APPRIS, a system for annotating principal coding isoforms, is a joint project between Centro Nacional de Investigaciones Oncologicas and Instituto Nacional de Bioinformatica. These are now available in BioMart.
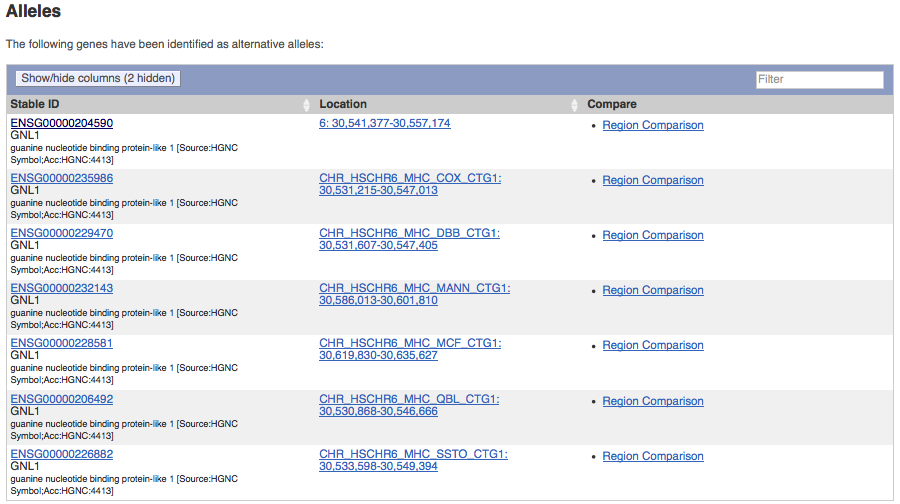
Finding gene alleles
The human and mouse genome assemblies comprise haplotypes (eg. MHC, LRC) and other alternate sequences where the same gene may be annotated in more than one of the alternate sequences. The gene summary page has now been improved to display all manually annotated alternative alleles for a gene. This is available from the left-hand side menu under ‘Gene alleles’.

Alternate alleles for GNL1 (ENSG00000206412) can be compared by clicking the Region Comparison link.
Interactive species tree
It is now possible to view the Ensembl species tree with our new interactive view. Once on the Species tree page, click the link “View the full Ensembl species tree”. You can select between Ensembl and NCBI taxonomy trees, and choose to view only a subset of species at a time.

Export of comparative genomics views
Export options for the comparative genomics views have undergone major changes in recent releases and in e78 we have finalised this work. The GeneTrees can now be exported in multiple formats. This option is available dirctly by the GeneTree image, replacing the ‘Gene tree (text)’ and ‘Gene tree (alignment)’ options. Orthologues and paralogues can be downloaded only in OrthXML format while gene gain/loss trees can be downloaded in PhyloXML format.

Available options for exporting GeneTrees.
Other news
- Updated pig HAVANA annotation in Vega.
- Updated C. elegans annotation (WS245).
- Updated data from the Animal QTL database for chicken, cow, horse, pig and sheep.
- RNASeq data can now be displayed as BigWig tracks, which will enable faster rendering of these data.
A complete list of the changes can be found on the Ensembl website.
Find out more at the Ensembl Release Webinar e78 (Wednesday, 17 Dec 4-4:30 GMT). Register here (for free!): http://tinyurl.com/e78-webinar.