If you’re trying to work out which variants are associated with a phenotype or disease, a major thing you might want to know is if someone else has already spotted it. And if not the variant, maybe the gene that it hits. You can get that through the VEP.
Phenotypes are available as a tickbox option in the online version of the VEP. If it is selected, phenotypes and diseases linked to either the gene or the variant will appear in the table. If there are more than three associations the table will initially just show you the number, but it can be expanded to show all the phenotypes, including their source and which colocated variant or gene they’ve been associated with. Since it shows you gene/phenotype associations, you can get potentially useful information even for novel variants.
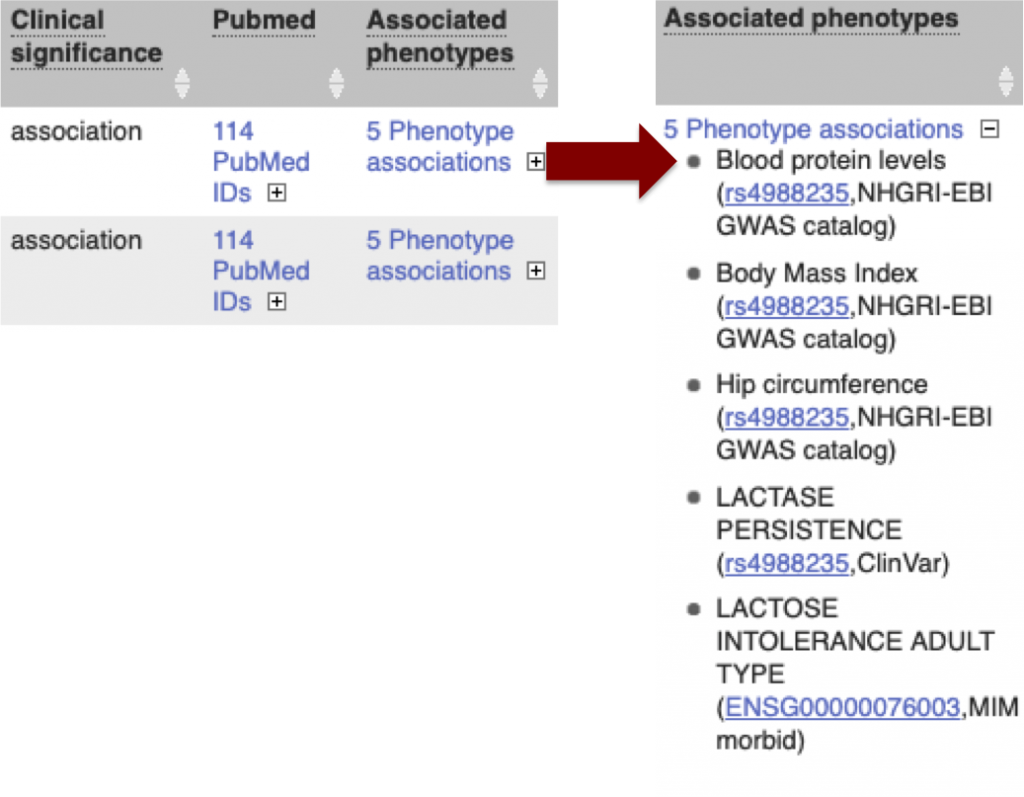
The associated phenotypes column shows the number of phenotypes linked to either the variant or the gene, and can be expanded to show a list
If you’re using the VEP endpoints on the REST API, phenotypes can be added using Phenotypes=1 in all the endpoints, including POST. To get phenotypes with the offline package, you’ll need to use the Phenotypes plugin, which works best with json output format.
Different types of data are available including statistical associations from the NHGRI-EBI GWAS Catalog and clinical assertions from ClinVar. At present only phenotypes linked to germline variants are shown in the web interface.
