If a variant hits a splice site, you want to know if splicing is going to occur as normal, or if you can expect a different protein isoform. We have a few cool tools with the VEP that will help you to assess that for your own variants.
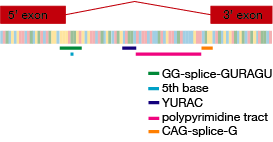
Functionally important bases highlighted in a splice junction
SpliceRegion was developed in collaboration with researchers from the DDD project, who examined splice site mutations in individuals with developmental disorders to identify loci of functional importance in splice sites. We were able to define two new splice consequences, splice_polypyrimidine_tract_variant and splice_donor_region_variant, which can be annotated on variants if you use the plugin with the VEP script. Their analysis is available as a pre-print.
GeneSplicer is another plugin that assigns a state to splice sites affected by variants (no_change, diff, gain or loss), followed by information about if this is a donor or acceptor splice site and a confidence assertion. This is only available with the VEP script.
Similar to missense pathogenicity predictors, splice site predictors score how likely it is that splicing will be changed by a variant, and if it will increase or decrease splicing at a particular site. Two scores are provided by the dbscSNV plugin along with related functional annotation, while the MaxEntScan plugin also provides a score of splice site entropy. Both of these plugins are available with the web interface, REST service and VEP script.
