We’ve just released Ensembl Genomes 94, which includes genomes for Emmer wheat and over 200 new fungi, updated gene trees and host-pathogen interactions from PHI-base.
New assemblies and gene annotation
Crops

The evolution of modern day bread wheat. Image courtesy of the IWGSC.
If you’re feeling hungry, you’ll be pleased to see two new crops in Ensembl Plants: Emmer wheat (Triticum dicoccoides) and Mung bean (Vigna radiata). Emmer is one of the precursors to bread wheat, having the AABB genome, which hybridised with goatgrass to give us modern wheat. It’s also an interesting plant in its own right, as it’s also used to make bread in certain regions, and is the wild relative of durum wheat, which is used to make pasta. Mung bean is used in Asian cookery such as dal in India, ice cream in Hong Kong and ginisáng monggó stew in the Philippines.
Fungi
We’ve done another automatic import of fungal genomes from the ENA, bringing in over 200 new species and strains like Puccinia coronata, a new and fast spreading pathogen of barley and oat. Alongside this, Saccharomyces cerevisiae, bakers and brewers yeast, and a major model organism for molecular biology, has an updated gene set, which will appear in both the main Ensembl site and Ensembl Fungi.
Comparing genes and genomes between species
Our gene tree pipeline, which is used to predict orthologues and paralogues, has been updated: we will be using HMMs to define our gene trees. The advantage of this is that we can now add new species and genes to the tree, without disrupting the existing genes. This is a great improvement on the old version, as it means that we won’t have significant changes between trees each release. The new pipeline has been run for vertebrates and plants, and will be run for other species sets in future.
We have also updated our pan-compara analysis, which constructs gene trees from representative species from all divisions of Ensembl. This includes updated gene annotation from Leishmania major, Physcomitrella patens, Chlamydomonas reinhardtii, Aspergillus nidulans, Saccharomyces cerevisiae and Danio rerio.
In other news, we’ve added approximately seventy new pairwise whole genome alignments between the genomes of crops and model organisms in Ensembl Plants. This includes alignments between all six of our legume genomes and the model legume, Medicago truncatula, hinting at some interesting biology. They have also enabled the gene gain/loss tree, giving you an overview of the pattern of gene duplication and loss over the course of evolutionary history.
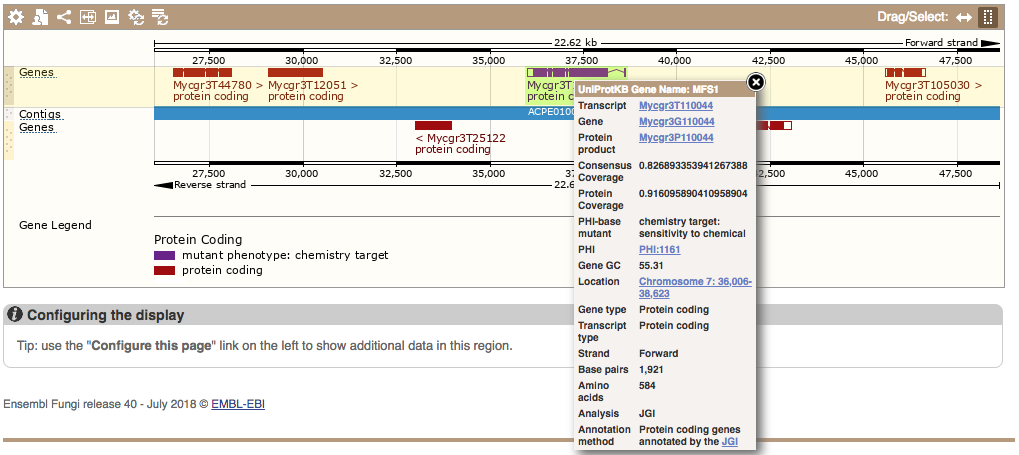
Host-pathogen interactions
We have refined our pipeline that imports data on host-pathogen interactions from PHI-base, improving the specificity. These data map PHI-base terms to genes, indicating how these genes are linked to phenotypes such as loss of pathogenicity or chemical sensitivity, which are involved in how the pathogen interacts with its host. We have PHI-base terms attached to genes in thousands of species in Ensembl Bacteria, Fungi and Protists. You can search for PHI-base terms, find them on genes and use them in BioMart.
Find out more
If you would like to find out more about these new changes, see live demos on how to find new data in the site, and ask questions to the Ensembl team, please register for the release webinar at 4pm (BST) on Tuesday 9th October. A recording of this webinar will be available on our YouTube channel.