One of the biggest highlights of the new Ensembl Plants release 40 is the inclusion of the new Wheat (RefSeq v1.0) genome from the International Wheat Genome Sequencing Consortium (IWGSC).
One of the biggest highlights of the new Ensembl Plants release 40 is the inclusion of the new Wheat (RefSeq v1.0) genome from the International Wheat Genome Sequencing Consortium (IWGSC).
The path to sequencing the wheat genome has been no easy ride, due to its large and highly repetitive genome. This new assembly from the IWGSC bridges many gaps from the initial genome sequencing effort. Read on to find out more about this exciting new genome assembly!
Wheat – the best thing since sliced bread
Why should I care about wheat?
Even if you’re not a fan of pizza, beer or cake you can’t deny that wheat is one of the world’s most important plants. We humans have been cultivating it since we left behind our nomadic lifestyle, and since then it’s spread its roots to become the most widely grown crop in the world. Wheat is a grass (part of the Poaceae family), and is particularly favoured for its high protein, fibre and essential nutrient content.
Research on wheat to improve yields, and increase resistance and tolerance to biotic and abiotic stress is critical, as current estimates predict that by the year 2050 we won’t have enough wheat to feed the population. This new genome assembly will enable the wheat research community to better understand wheat genetics and be able to better tackle such challenges.
The first wheat genome assembly
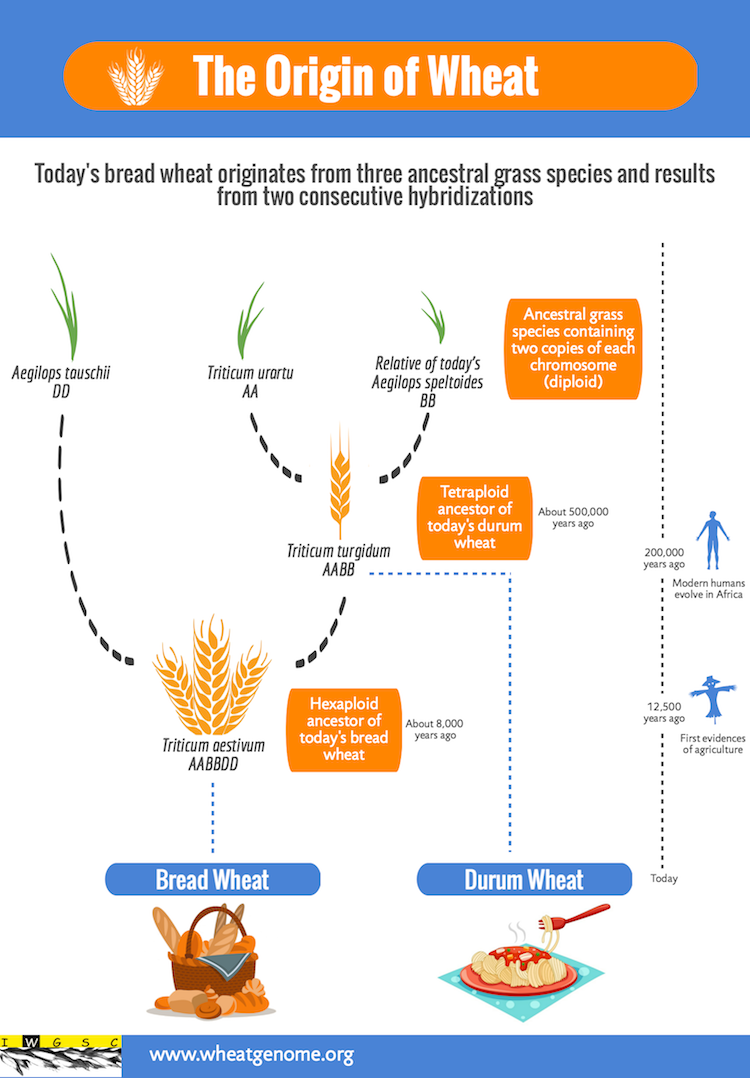
The popularity of wheat is not the only thing that has grown over time – so has it’s genome! The original wheat species grown was Einkorn wheat (Triticum monococcum), which was a modest diploid organism, with only seven chromosomes. Multiple spontaneous hybridisations have developed modern bread wheat (Triticum aestivum), which is a hexaploid meaning it has six copies of each of its seven chromosomes (Figure 2).
The large genome size and the highly repetitive nature of the sequences led to delays in the initial attempt to sequence the wheat genome, where research shifted to focus on creating a database for the conserved coding sequences that make up about 2% of the genome, published in 2009.
The first draft wheat genome sequenced by the IWGSC was released in 2014, with sequences from Chinese spring wheat. The final sequence length was 17,000,000,000 base pairs (17-gigabases) – to put it in perspective this is about five times larger than the human genome!
Wheat genome assemblies in Ensembl Plants
The Ensembl Genomes site, including Bacteria, Fungi, Metazoa, Plants and Protists was first launched in 2009, eight years after the Ensembl main site for vertebrate genomes. Wheat was first introduced to the site in 2014, with the first IWGSC assembly (IWGSC v1.0). Shortly after this we updated the assembly to the Earlham Institute’s (previously The Genome Analysis Centre (TGAC)) 2015 assembly annotation. Ensembl now includes the newest wheat genome assembly, from the IWGSC (IWGSC RefSeq v1.0 and IWGSC RefSeq annotation v1.0).
This new assembly version not only brings updated gene and transcript annotation from the IWGSC, but also presents other novel features from other external databases that we’ve aligned to this new assembly.
These include:
- Axiom 820K SNP Array from CerealsDB.
- EMS-induced mutations from sequenced TILLING populations (Kronos and Cadenza).
- Variant consequence predictions from the Sorting Intolerant From Tolerant (SIFT) algorithm.
- Assembly to assembly mapping and gene ID mapping to the previous TGAC v1 assembly and annotation.
- Whole genome alignments to rice (Oryza sativa Japonica IRGSP-1.0), brachypodium (Brachypodium distachyon v3.0) and barley (Hordeum vulgare Hv_IBSC_PGSB_v2).
- You will also be able to use our polyploid viewer on the new wheat genome assembly, so you can compare regions across all three components (A, B, D – see Figure 3) simultaneously.

Figure 3: Example genomic region showing the wheat polyploid view
If you have been working on the previous wheat genome assemblies, and find that your data has changed with the new coordinate system, don’t worry! You can map your coordinates from the older assemblies to the newer version with our Assembly Converter tool. We will still keep the TGAC v1 assembly accessible through the Ensembl Genomes archive sites if you want to revisit this.