What’s New in e85:
- 30 new human epigenomes from the Roadmap Epigenomics Project
- Human and mouse: Updated GENCODE set; including manually annotated HAVANA annotation, and all CCDS genes
- Imported symbol names from the Vertebrate Gene Nomenclature Committee (VGNC) for Chimpanzee
- Improved highlighting options in the Location View for Userdata and Tracks
- Wasabi Tree viewer
30 New Epigenomes from the Roadmap Epigenomics Project
Roadmap Epigenomics is producing epigenomic maps for stem cells and primary ex vivo tissues selected to represent the normal counterparts of tissues and organ systems frequently involved in human disease. In Ensembl 85, we have run our regulatory pipeline on Roadmap Epigenomics data for 30 cell/tissue types.
Additionally, the peak calling component of the Ensembl Regulation Sequencing Analysis pipeline has been improved. All of the existing ENCODE and BLUEPRINT data in Ensembl’s Regulation database have been reprocessed.
Human Gene Set Update and New Assembly Patches
The human gene set now corresponds to GENCODE 25 and the assembly has been updated to include new assembly patches for GRCH38.p7
VGNC Symbols for Chimpanzee
We now import symbol names from the Vertebrate Gene Nomenclature Committee (VGNC) for Chimpanzee. VGNC is an extension of HGNC for standardising naming across vertebrates lacking a nomenclature committee by transferring gene symbols from human to known orthologues. This replaces our own system for naming chimpanzee genes. This has improved our naming of chimpanzee genes as seen in POMGNT2, which was previously named GTDC2 in Ensembl 84.
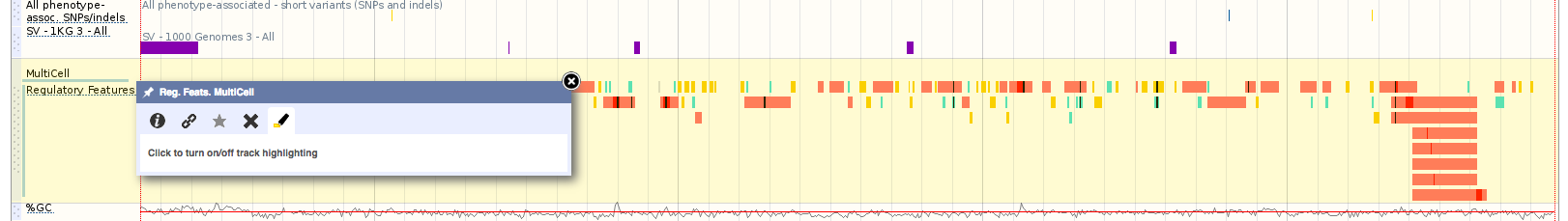
Userdata and Track Highlighting
We have updated the highlighting functions in Location View:
- User data Highlighting – When you upload your own data to Ensembl the newly uploaded track will be automatically highlighted. This highlighting will disappear when you hover your cursor over the track
- Highlight track on hover – When placing the cursor over any track, the whole width of the track will be highlighted
- Track menu Highlight Icon – Track menu now contains an icon that allows you to manually turn highlighting on/off
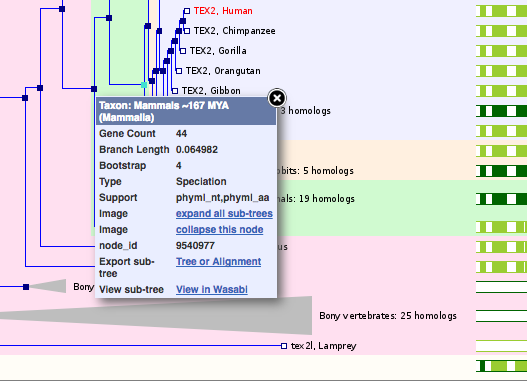
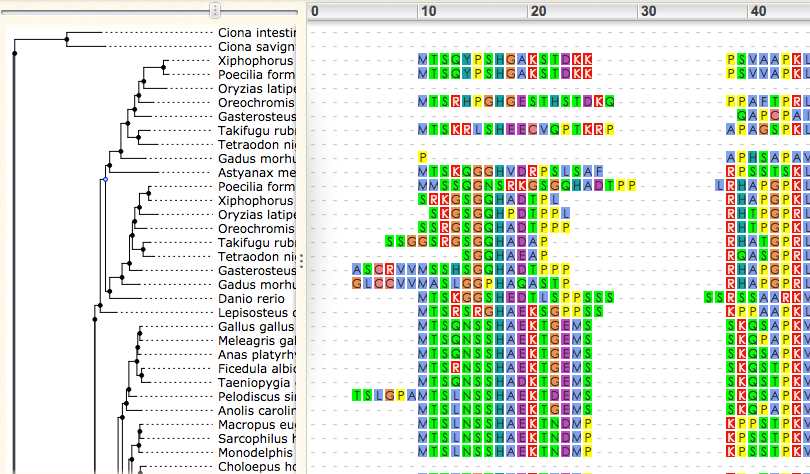
Wasabi Tree Viewer
Wasabi has replaced Jalview as a way to view gene trees and multiple alignments. Clicking on any node within the gene tree will give you the option to ‘View in Wasabi’.
Clicking on ‘View in Wasabi’ will open a pop-up window with the tree and alignment:
Other News
- Updated Ensembl-Havana rat gene set; a merge of complete Ensembl gene models and the latest Havana gene annotation
- Human and mouse CRISPR sites, predicted by Wellcome Trust Sanger Institute Genome Editing (WGE) have been added
- Human and mouse databases have been updated to dbSNP147 and dbSNP146 respectively
- Phenotype data updated for several species, including human, mouse, pig and chicken
- GTEX eQTL data for 14 human tissues has been added to the Gene Regulation view
- The Allele Frequency Calculator has been migrated from the 1000Genomes website to our GRCh37 archive. This tool takes a VCF file and a matching sample panel file, and calculates allele frequencies for one or more 1000G populations for a defined chromosomal region
-
New tool: File Chameleon customises files from the Ensembl FTP server. Current functions include; adding ‘chr’ to your chromosome names for use with UCSC’s genome browser and removing long genes
A complete list of the changes can be found on the Ensembl website.
Find out more about the new release and ask the team questions, in our free webinar: Wednesday 27th July, 4pm BST. Register here.