Continued HapMap variation data access through Ensembl
NCBI have recently released plans to immediately retire their HapMap interface, however, data from the HapMap Project will continue to be freely accessible through Ensembl. There is lots of help and documentation as well as video tutorials to help you learn how to access variant data in Ensembl. This post aims to complement those materials to highlight the methods for accessing the HapMap Project variant data specifically.
Finding HapMap variants by ID
You can find data from the HapMap project relating to specific variants by searching for the variant rsID itself. In Ensembl, you can find information related to variants identified in the HapMap Project, which includes population genetics statistics:
However, as you can see from the example above, some of the populations represented in the HapMap Project have two separate entries in the Population Genetics table. This is because the HapMap Project was completed in a number of phases. In the first phase, a number of different groups used different genotyping platforms to type variants from a number of population panels (CEU, YRI, HCB, JPT). In a later phase, a larger set of samples were added to the samples from the initial phase and submitted as HapMap3. The two entries refer to the two submitted phases of the HapMap Project, where the number in brackets next to the allele frequency indicates the number of samples in that population.
It is also possible to view HapMap Project results by gene of interest by searching the Variant Table. The Variant Table can be filtered by ‘Evidence’ type so you can choose to see only HapMap Project variants, for example.
Filtering the Variant Table by ‘Evidence type = HapMap’ will filter the displayed variants to those identified in the HapMap project. This will be denoted by the![]() in the Evidence column.
in the Evidence column.
Finding HapMap Project variant data using BioMart
HapMap SNP data can also be retrieved using BioMart. There is help and documentation and a video tutorial to help you while using BioMart.
When querying the Homo sapiens short variants dataset in BioMart, you can access HapMap variant data specifically by using the ‘Variant Set Name’ filter and selecting the HapMap populations that are relevant for your research.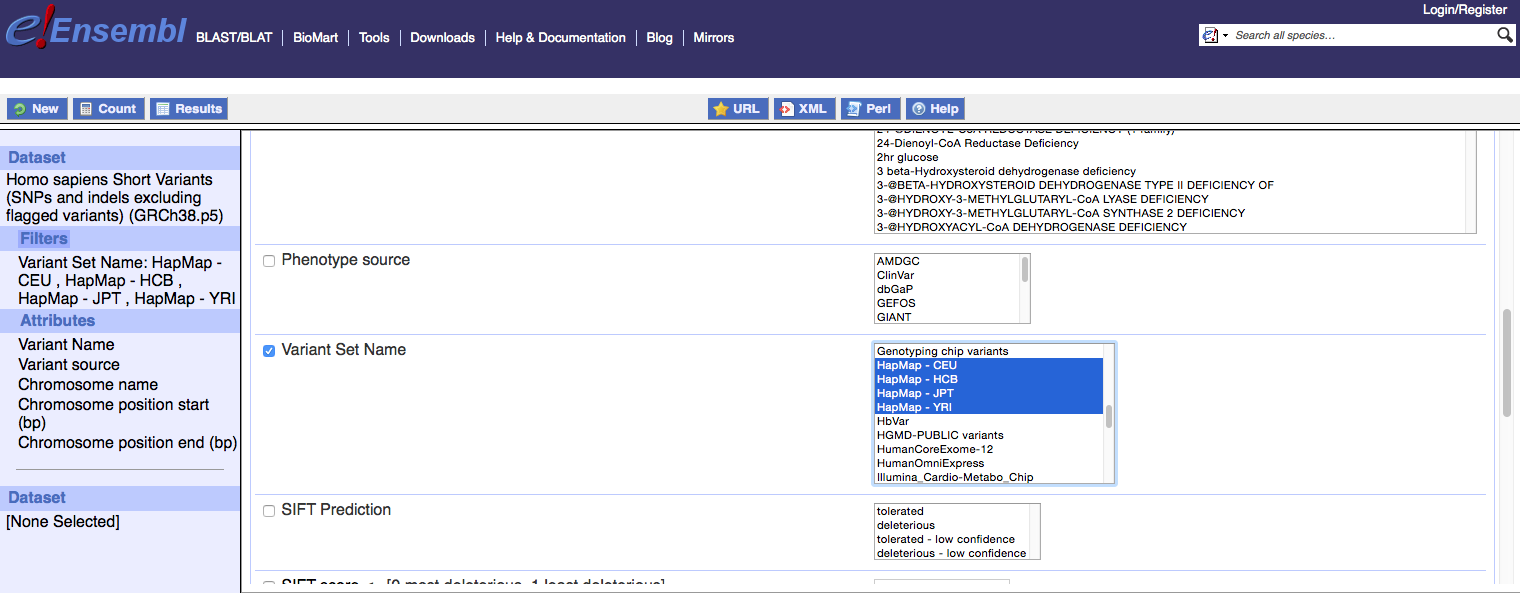
Finding HapMap variants using the Ensembl API
It is also possible to access variation data through the Ensembl APIs. Using the Perl API, for example, it is possible to retrieve variation data specifically related to the HapMap Project variant set, either as the whole HapMap variant set, or as individual populations represented in the HapMap Project.
