What’s new in e84:
- Human: Incorporation of BLUEPRINT Epigenome data and methylation data
- Pairwise Linkage Disequilibrium (LD) calculation on LD variant page
- Track hub registry interface
- Transcript haplotype view
Incorporation of BLUEPRINT Epigenome data
BLUEPRINT is a large scale research project aimed at deciphering the epigenome of blood cells. ChIP-seq and DNase hypersensitivity data from the BLUEPRINT project has now been incorporated into Ensembl. All of the cell types analysed in the BLUEPRINT project are listed here. In Ensembl 84, we are including BLUEPRINT data for the following 20 independent cell types, divided based on cell lineage and tissue source:
CD14+ CD16- monocyte from Venous Blood
CD14+ CD16- monocyte from Cord Blood
CD4+ ab T cell from Venous Blood
CD8+ ab T cell from Cord Blood
CM CD4+ ab T cell from Venous Blood
eosinophil from Venous Blood
EPC from Venous Blood
erythroblast from Cord Blood
HUVEC prol from Cord Blood
M0 macrophage from Cord Blood
M0 macrophage from Venous Blood
M1 macrophage from Cord Blood
M1 macrophage from Venous Blood
M2 macrophage from Cord Blood
M2 macrophage from Venous Blood
MSC from Venous Blood
naive B cell from Venous Blood
neutro myelocyte from Bone Marrow
neutrophil from Cord Blood
neutrophil from Venous Blood
This data can be viewed alongside other tracks in Ensembl by using the ‘Configure this Page’ option and selecting your cells of interest. ![]()

Pairwise LD calculation
You are now able to calculate linkage disequilibrium (LD) between any two variants in Ensembl. To calculate the r2 and D’ values for LD between two specific variants, enter the ID of any variant into the LD calculation text box on the specific page of the reference variant. This feature can be found by clicking on ‘Linkage Disequilibrium’ from the menu on any variant page.
Track Hub registry interface
With the arrival of the new Track Hub Registry, we have added a feature that allows you to search for track hubs of interest and attach them directly to Ensembl. Just click on the ‘Add your data/Manage your data’ button on any Ensembl page, and select ‘Track Hub Registry Search’ from the lefthand menu. ![]()

The interface will only search for hubs that have assemblies available for the site you are on; to see the full range of species and assemblies, visit the Track Hub Registry site directly.
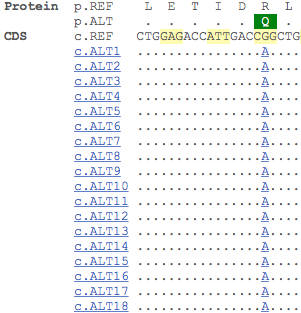
Transcript haplotype view
The transcript haplotype view is a new data view we have implemented that allows you to explore observed transcript sequences that results from variants identified from resequencing data from the 1000 Genomes Project. By clicking on the ‘Haplotypes’ link on any transcript page, you are able to view protein consequences, population frequencies and protein alignments of all the haplotypes for that particular transcript.
Other news
- Mouse: update to GENCODE M9 annotation
- Zebrafish: updated gene set, including manually annotated HAVANA annotation
- Baboon: lincRNA model update
- Latest sequence variants from dbSNP build 146 for human, cow and dog
- Import of COSMIC 75 cancer data
- New and updated studies from DGVa for several species such as human, mouse, zebrafish, macaque, cow and dog
- Gene trees: new option to prune by target species/ taxon in the REST API
- Ensembl Families now defined by an HMM library, based upon the Panther database.
- Alignments in CRAM format
- DAS support ended
- Regulatory segments retired from the Ensembl regulation BioMart, but now available in bigbed format through the ftp site
A complete list of the changes can be found on the Ensembl website.
Find out more about the new release, and ask the team questions, in our free webinar. Wednesday 16th March, 4pm GMT. Register here.