What’s new in e82:
- Ensembl mobile website
- Support of VEP Plugins through the web interface, script and REST
- Improved Variation tables
- RNASeq data set of Zebrafish developmental stages
- Marking a region on images
- LastZ replaced TBlat for pairwise alignments
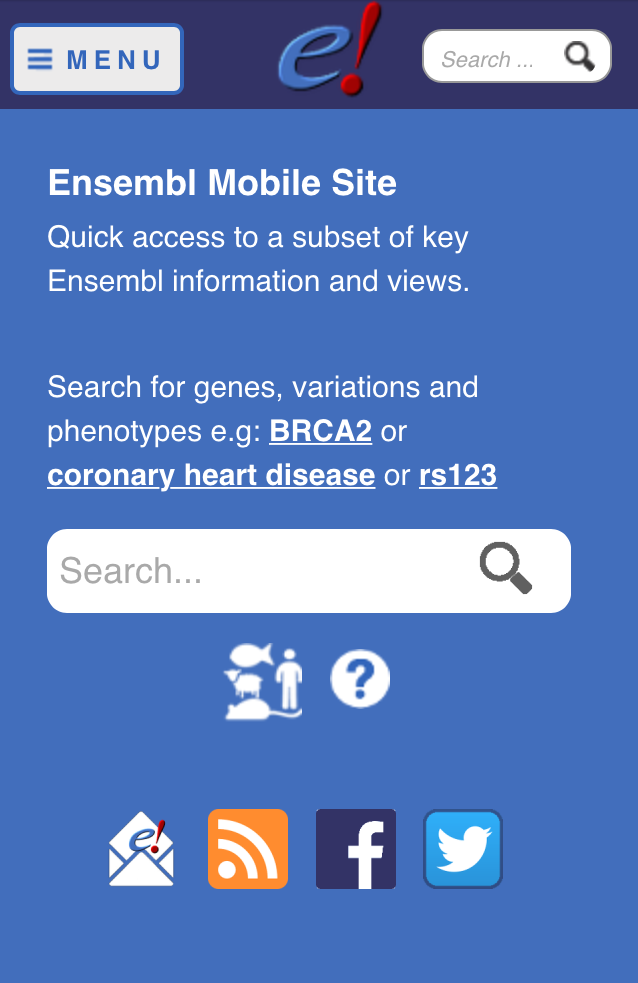
Ensembl mobile website
We are very happy to announce the release of the Ensembl website mobile version, available on http://m.ensembl.org. This new website allows you to quickly search for a gene, variants or phenotype on your mobile device.
Support of VEP Plugins through the web interface, script and REST
The VEP can now be extended beyond its core functionality using a system of plugins. Plugins are a powerful way to extend, filter and manipulate the output of the VEP. More information regarding the VEP plugins can be found on the following documentation page.
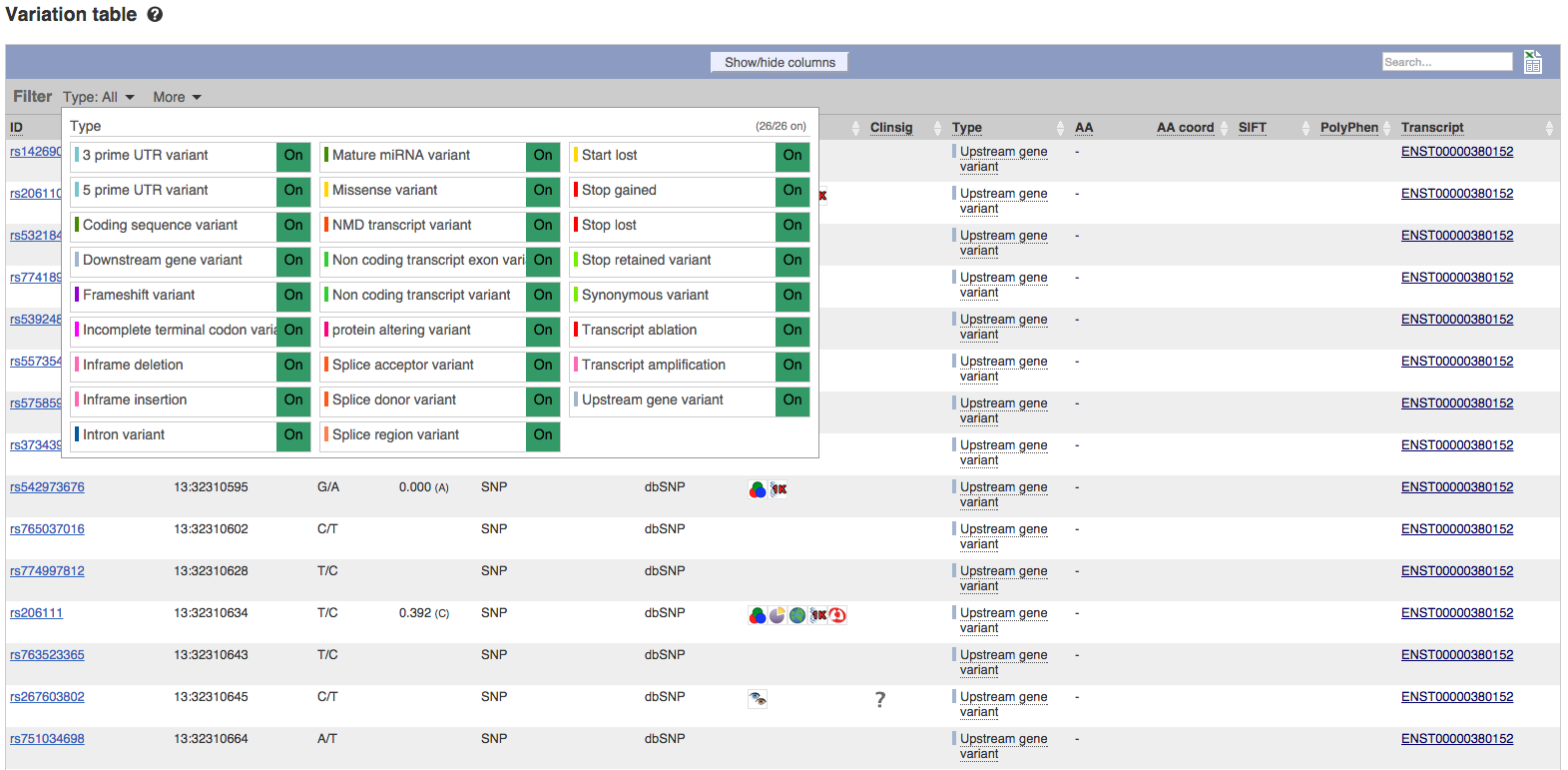
Improved Variation tables
Variation tables for genes and transcripts have been reimplemented to effectively handle the large number of variants now known for many genes. At the same time, the ability to filter, sort, and select this data has been improved. Filtering by variant type is now achieved by selecting the “Type:” filter at the top of the main table. Further features and refinements are expected to be added in forthcoming releases.
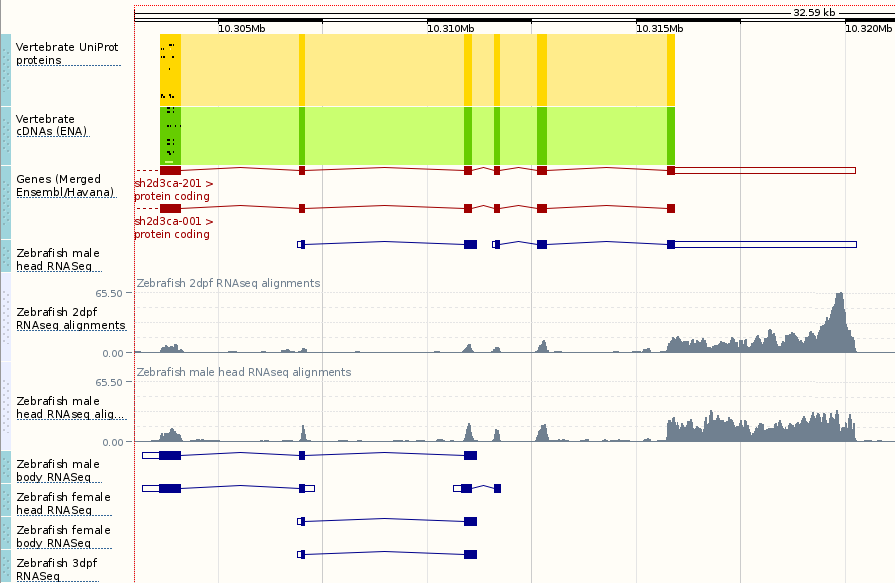
Zebrafish development stage RNASeq data set
We’ve added sample-specific BAM files, splice junctions (introns) and gene models based on a range of zebrafish developmental stages and tissue samples.
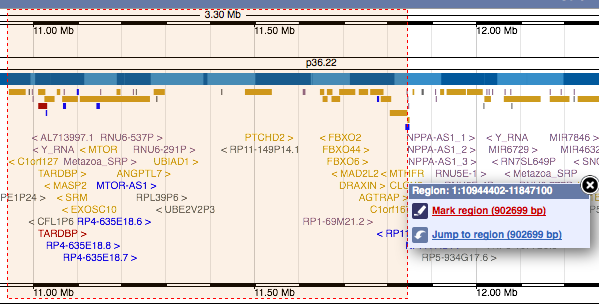
Marking a region on images
A new feature to mark a selected region has been added to the location, gene and other views. Marking can be applied by drag-selecting a region and then using the zmenu to mark it, or by clicking on a feature on an image and then using the zmenu to mark the location of the feature.
LastZ replaced TBlat for pairwise alignments
We have replaced TBlat with LastZ and recomputed 9 pairwise alignments using LastZ. TBlat was used for distantly related species as it was yielding a higher genome coverage, but over time we have optimised the LastZ parameters that enable it to give a 50-100% increase in genome coverage.
Other news
- Human variation data updates to dbSNP (144) including variants from the Exome Aggregation Consortium (ExAC)
- Mouse: updated to GENCODE M7 including HAVANA annotation.
- Improved data upload form
- Improvements to PDF export
- Export mode for projectors and print
- Phenotype data updated for several species, including human, mouse, rat and horse
A complete list of the changes can be found on the Ensembl website.
Find out more about the new release, and ask the team questions, in our free webinar. Wednesday 7th October, 4pm BST. Register for free.