From release 68, we are using Sequence Ontology (SO) terms for the variation consequences, in an effort to standardise terms across the different browsers, making it easier for users to do a cross comparison of variation annotation. The UCSC Genome Browser will use these terms on their SNP details page around mid-August, dbSNP will update their web display in the next few weeks and the ICGC also intend to standardise on SO terms for describing somatic mutation consequences.
At the same time, we have added a couple more specific consequences for SNPs and in-dels (splice donor variant and splice acceptor variant for example) and consequences for larger structural variants are now available through the Variant Effect Predictor (VEP). The complete list of terms and definitions are in our documentation.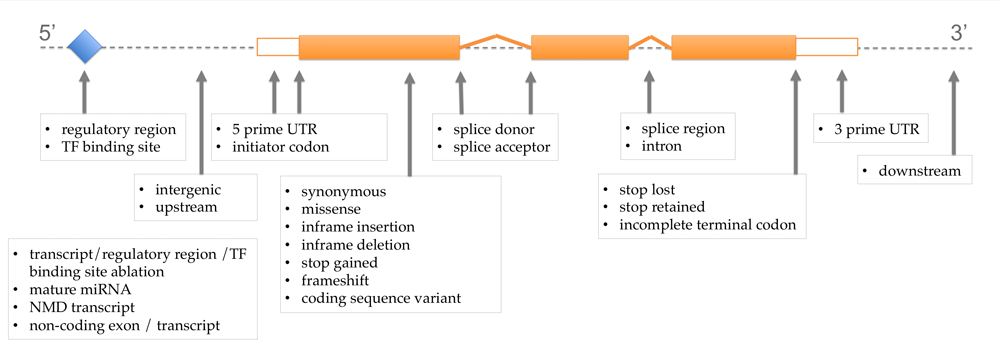 As you will see, the SO equivalents for our old terms are fairly straightforward. The most notable difference is that we have replaced “non-synonymous” with the more specific term “missense”, for changes in amino acid which do not include stop gained, as we already have a specific term for stop gained.
As you will see, the SO equivalents for our old terms are fairly straightforward. The most notable difference is that we have replaced “non-synonymous” with the more specific term “missense”, for changes in amino acid which do not include stop gained, as we already have a specific term for stop gained.
![]() The old Ensembl terms are still available on the website (using”Configure this page”) and if you have text files or VEP output files with our old Ensembl terms, you can easily update these to using the SO terms by running the following script.
The old Ensembl terms are still available on the website (using”Configure this page”) and if you have text files or VEP output files with our old Ensembl terms, you can easily update these to using the SO terms by running the following script.
